THERAPEUTIC VULNERABILITIES
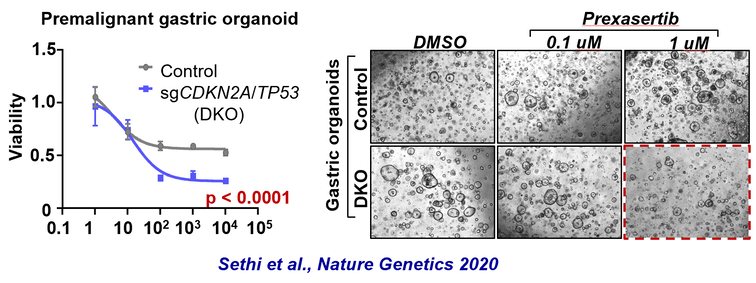
Using the integrative mouse model, we demonstrated that select genomic subtypes of gastric and esophageal cancer are susceptible to DNA damage response (DDR) inhibitors. Based on these results, we are pursing preclinical testing of DDR pathway inhibitors to investigate the therapeutic efficacy of these agents alone and in combination with specific chemotherapeutic agents.
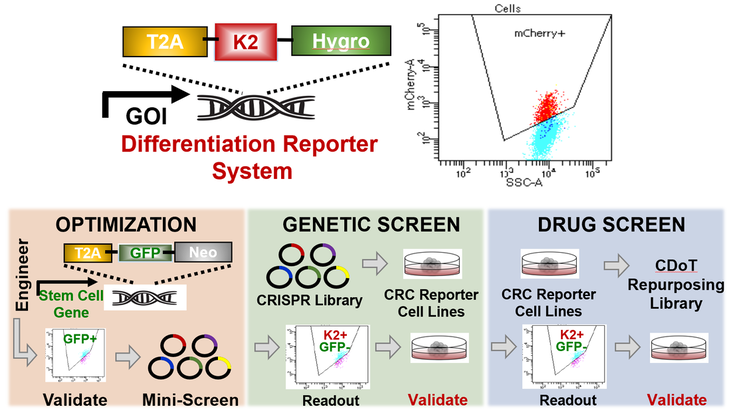
Using a multidisciplinary approach, we demonstrated that hyperactive stem cell programs block proper cell maturation in colorectal cancer (Liang et al., Gastroenterology 2022). Taking advantage of this understanding, we have designed a new endogenous reporter system that measures stem cell and differentiation activity using different fluorescent probes. With this platform, we will perform genetic screens to determine the key factors that drive hyperactive stem cell signaling and differentiation blocks. Furthermore, we will use this system to discovery drugs that overcome differentiation blocks in colorectal cancer. We strongly believe that the next generation of therapeutics in the treatment of colorectal cancer will be directed at restoring proper differentiation.